About Genomon
DNA sequence analysis pipeline
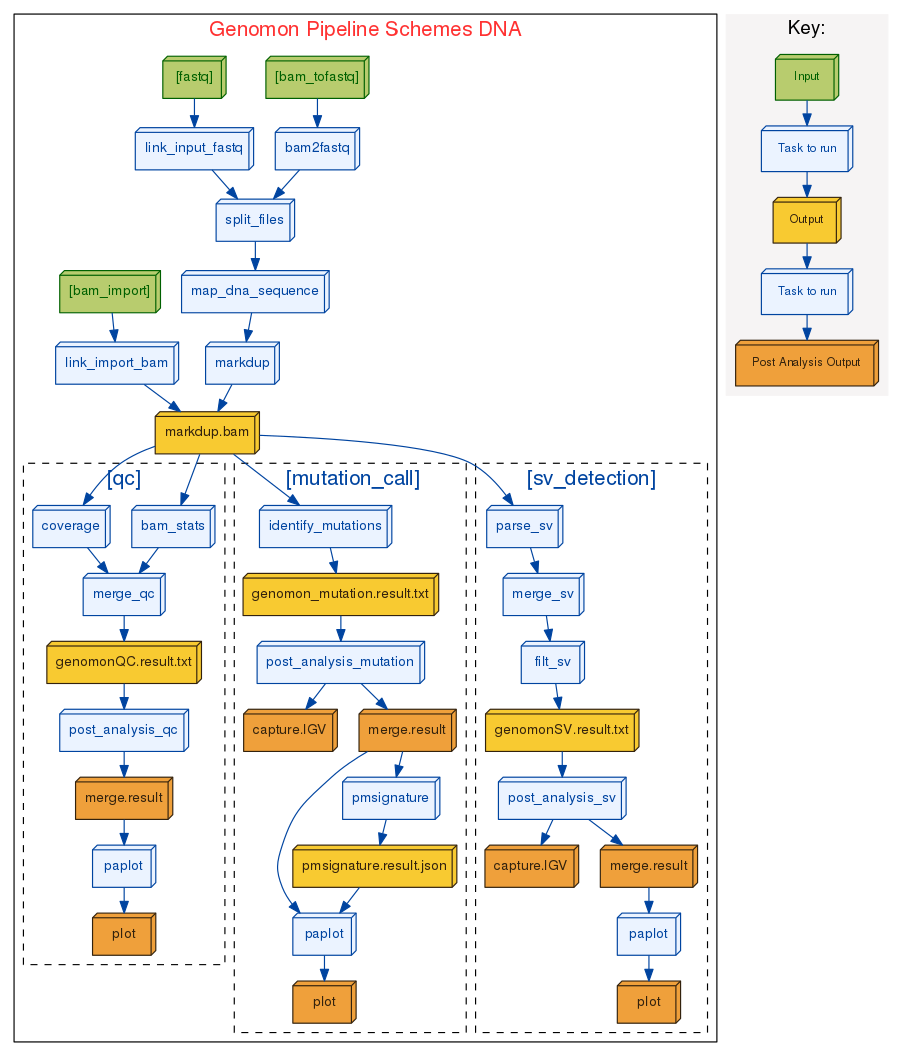
- Can start from both bam and fastq files.
- Suitable for whole genome sequencing analysis (as well as exome sequencing analysis).
- Carefully devised mutation filtering steps enables sensitive and accurate somatic mutation detection.
- GenomonSV detects mid-range indels (30bp - 300bp) such as FLT3-ITD as well as long range structural variations.
RNA sequence analysis pipeline
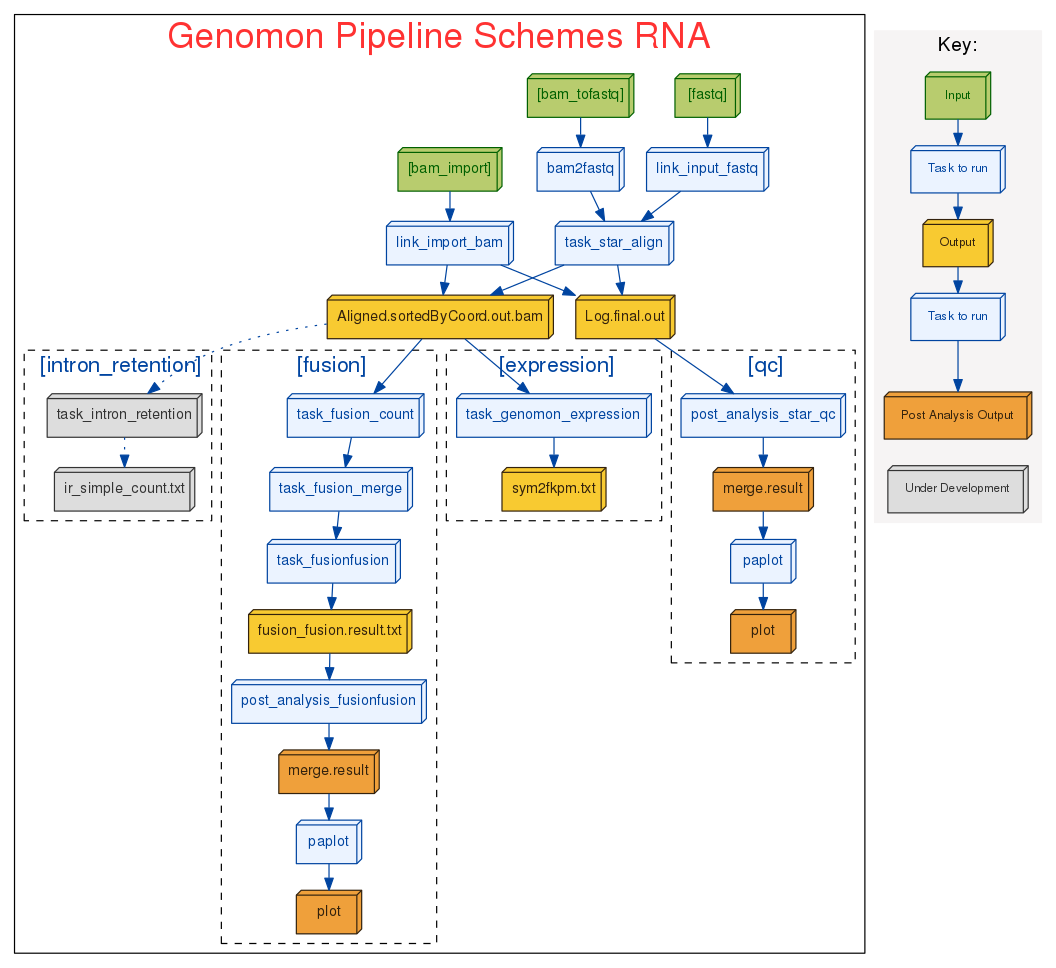
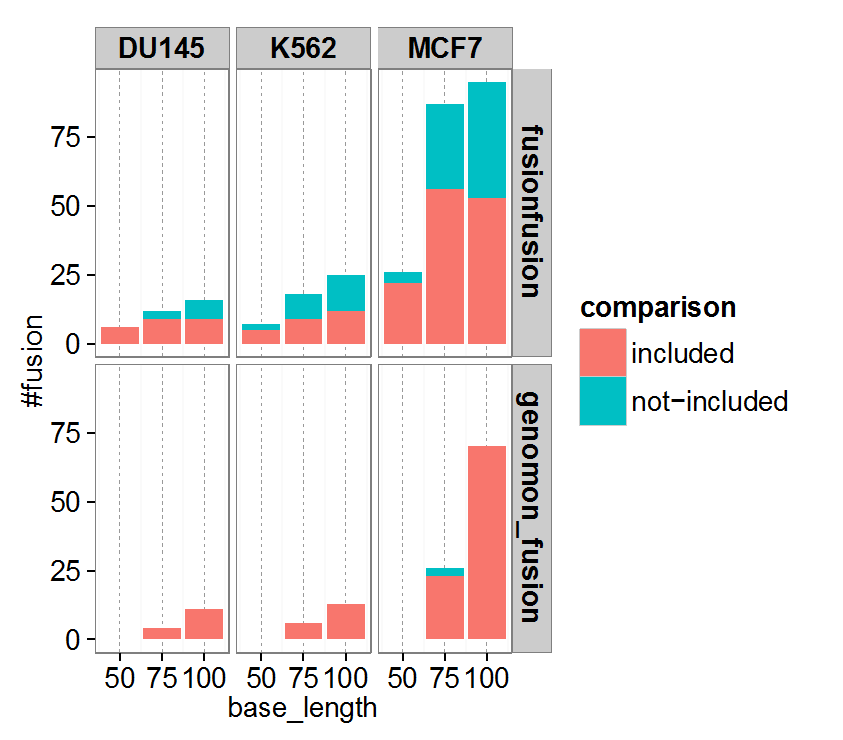
We renovated gene fusion detection (from Genomon-Fusion) to a new approach fusionfusion
- Filtering chimeric read generated by STAR.
- Computationally far more efficient compared to Genomon-Fusion (~200 holds)
- Sensitive even for short read length (~50bp).
- Can easily change to other alignment tools (such as map-splice2, tophat2) by option.
- Integrative analysis with DNA and RNA sequence data (e.g., detecting somatic mutations causing splicing changes) is coming out soon!!
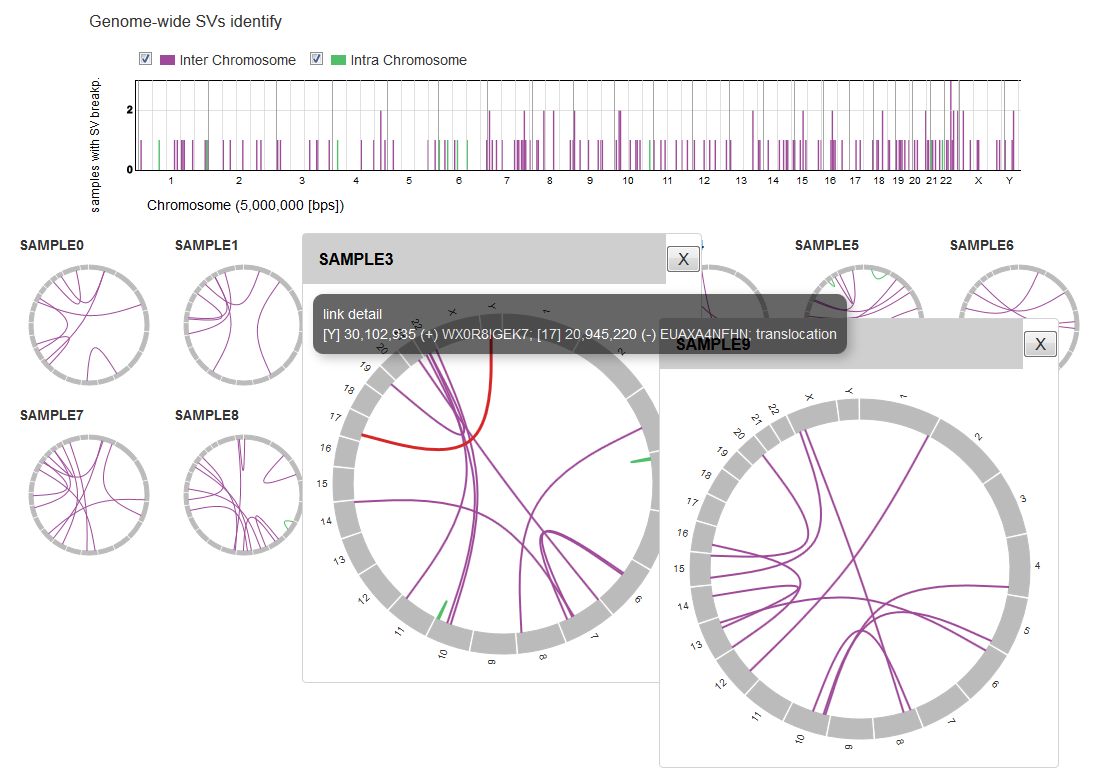
Automatic construction of interactice analysis graphs.
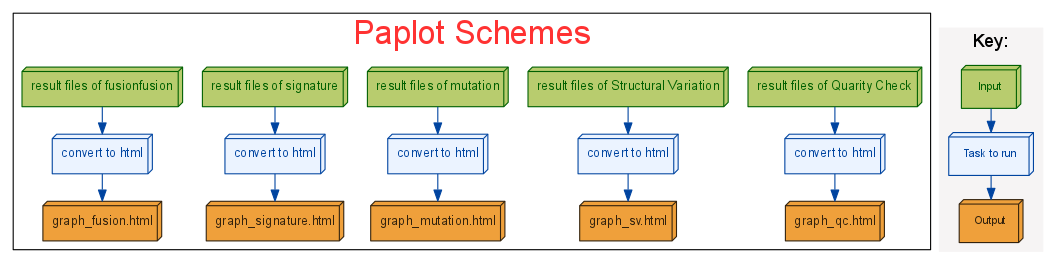
- Graphs are able to be sorted, selected.
- Extract some samples of your interest.